For this third installment, we introduce the highly coveted ability to edit salt data for batches, and a simplified workflow for registering new batches in registration vaults. Other enhancements include a standard deviation calculation for averaged data, as well as a black and white printing option for structure images.
Simplified Batch Registration (For vaults that use CDD's molecular registration system)
You can now manually register a new batch of an existing molecule without the need to enter the structure. The new batch inherits the core structure from the molecule record, while you can specify the salt and hydrate information. To add a new batch, go to the Batches tab of the molecule page and click "Add a Batch".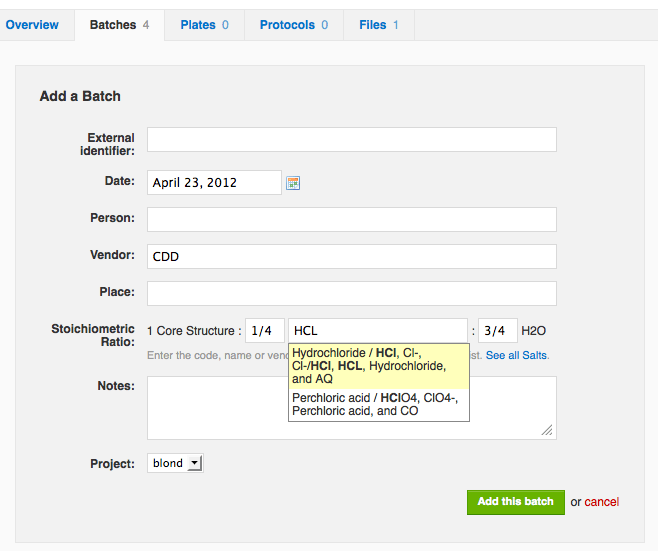
Editing Salt Information for Existing Batches (For vaults that use CDD's molecular registration system)
You may now edit salt data for existing batches in registration system vaults. Simply go to the Batches tab of the Molecule Show page, and choose to edit an existing batch. Update the Stoichiometric Ratio field following the same guidelines as for new batch creation, and formula weight and batch name automatically update to reflect the new stoichiometry and salt name.Molecule and Batch Creation While Manually Adding Readouts (For vaults that do NOT use CDD's molecular registration system)
To prevent errors during manual protocol readout entry, only existing molecules and batches can be updated with assay results. New molecules and batches must be explicitly registered during a batch data import via file, or manually using “Create New Molecule” and “Add a Batch” functionality, and can no longer be registered during protocol data entry.Standard Deviation and n for averaged readouts
When you set up an average calculation for a protocol readout definition, the standard deviation and the total number of points (n) are calculated and displayed alongside the averaged result. Results can be exported in the same format: Avg ± StDev (n=#)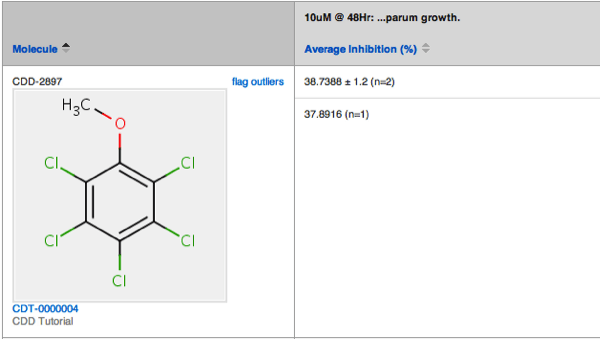
Export Structure Images in Black and White
If you have ever printed out structures exported from CDD, you may have noticed that atom color-coding does not print well on monochrome printers. Now you have the option to export structure images in black and white for better rendering. The option is found in the "Export Results" interface for Excel files.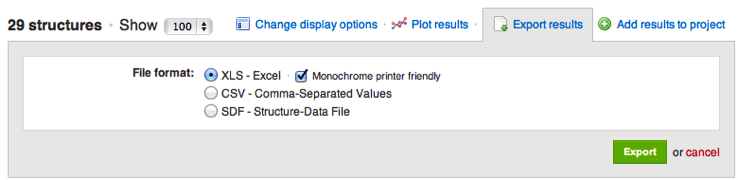
Other posts you might be interested in
View All Posts
Events
2 min
April 22, 2024
Recorded Presentations: CDD 20th Anniversary User Group Meeting
Read More
CDD Blog
9 min
April 19, 2024
Drug Discovery Industry Roundup with Barry Bunin — April 19, 2024
Read More
CDD Blog
2 min
April 19, 2024
CDD Appoints Yasushi Hamagashira as Head of Sales and Marketing for Japan
Read More


