A novel collaborative, fast-paced workflow: From a discussion on Twitter to a micropublication on Ebola
From the desk of CDD CSO Sean Ekins, M.Sc., Ph.D., D.Sc.
We are all aware of the nightmare going on in Africa, and now elsewhere, regarding the spread of the Ebola virus. Although it is important to point out so far it has killed a small fraction of the hundreds of thousands that have likely died in the same period from Malaria and Tuberculosis, it remains a deadly, infectious threat with limited available treatment options. However, at CDD, we strongly believe in the power of mining drug discovery data to hasten the development of new therapeutics. We have used this approach for many different infectious and neglected diseases, such as Malaria and Tuberculousis. Thus, we wondered if we could use a similar approach to attack the Ebola virus. In particular, I was interested in if FDA-approved drugs could be repurposed to treat Ebola. Indeed, this seems like a very promising approach and researchers from two separate groups have screened FDA approved compounds for activity against Ebola.
My exploration of this topic also offers an interesting vignette into how all sorts of new technology tools are quickening the pace of research. I started following Ebola several months ago, tracking the topic on twitter and RSS feeds using the ODDT mobile app:
Yesterday I found a paper by one of our collaborators Dr. Peter Madrid and his colleagues at SRI International. I was surprised to see he found many hits versus Ebola including the drugs chloroquine and amodiaquine. Both of these are widely used antimalarials. I posted a tweet on this:
Then one follower V.S. Schulz pointed me to a blog post from Derek Lowe, so I posted my comment there.
Then an American Doctor in Haiti (Megan Coffee) responded:
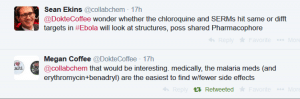
This paper by Johansen et al described 2 selective estrogen receptor modulators that were active in vitro and in vivo. I then speculated on a common pharmacophore for these 2 classes of drugs:
I decided to download all 4 structures from the ChemSpider mobile app, saved them in the mobile molecular datasheet App then Tweeted them. The ODDT app can deal with the molecule structures from the tweet and they appear as a thumbnail in the app.
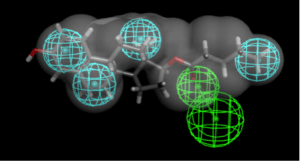
My next step was to build a common feature pharmacophore in Discovery Studio. This took 32 seconds to produce a pharmacophore with 4 hydrophobic features and a hydrogen bond acceptor. It was followed by searching a couple of datasets with previously computed 3D conformations (including a set of FDA approved drugs from CDD Public which contains over 2000 compounds). I found that adding the van der Waals shape of amodiaquine helped to limit the number of hits to about 120 in each case. These searches retrieved a few of the many hits from the Madrid et al., paper and in particular estradiol was a good fit. This is interesting as the molecule was not a part of the training set.
As I was doing this I put together a 1 page summary, a “micropublication”. I posted this document on figshare along with the results of the database searches and ultimately the original pharmacophore.
This represents a fascinating example of a workflow where a conversation on Twitter lead to an exploration of published repurposing data, possibly revealing a previously unpublished relationship between the molecules. They may also reveal that there is a target in Ebola that has a binding site that can deal with structurally diverse hydrophobic molecules and it would not surprise me if other steroids could also be explored that would overlap with estradiol. Of course one could also perhaps suggest that the antimalarials might be useful as SERMs and the SERMs might be useful antimalarials also. All these questions could be tested in future. In the mean-time there is an urgent need to find a treatment to cure Ebola. The work of Madrid et al., and Johanson et al., has pointed to FDA approved drugs which appear promising. I'd like to know if anyone at the CDC, NIH, NCATS or elsewhere is fast tracking this work into the clinic.
This blog is authored by members of the CDD Vault community. CDD Vault is a hosted drug discovery informatics platform that securely manages both private and external biological and chemical data. It provides core functionality including chemical registration, structure activity relationship, chemical inventory, and electronic lab notebook capabilities!
CDD Vault: Drug Discovery Informatics your whole project team will embrace!