It’s time to start writing about the BioAssay Express project, since it has been technically developed out in the open. This is what has been taking up the majority of Alex's time for the last half year or so, and it has the potential to make an important contribution to the drug discovery process.
First some background. In 2014, Collaborative Drug Discovery developed a way to use natural language processing & machine learning to guide human curators toward quickly and correctly annotating their text assays using semantic terms from the BioAssay Ontology. The proof of concept worked just great, and we published the results. As we planned the level-up to a real actual service, we quickly realized that the most troublesome rate limiting step was figuring out how to apply the available semantic terminology to an assay (i.e. creating good training data): it’s not a trivial problem, and nobody had worked it out, so we spent much time designing a schema for specifying how to use the available specialized terms to describe the bioassay protocols that we are focusing on. You can refer to the literature for a detailed account.
With these preliminary algorithms, datastructures and vocabularies, we were ready to embark on the next step: selecting a large collection of bioassay protocols with their procedures conveniently available. The go-to place for such data is of course PubChem, which provides over a million assay protocols. While most of these are of limited use for our purposes, it is relatively straightforward to select just the assays from the Molecular Libraries programme, which are almost all very well specified, with quite a lot of detail. There are thousands of them, which makes for a respectable training set.
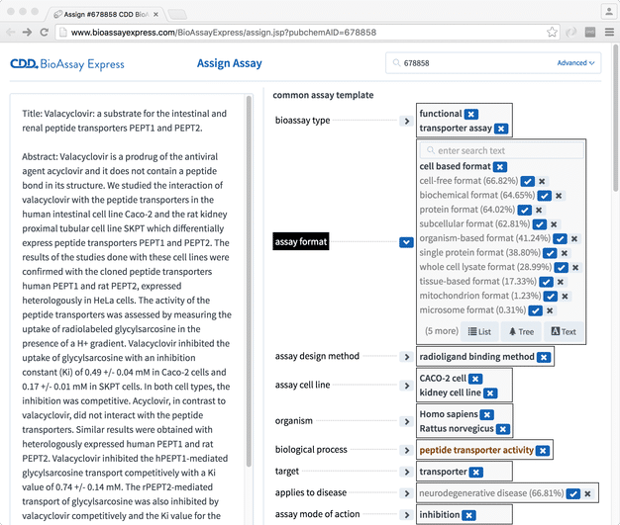
With the assays themselves compiled, our next task was to create a web interface so that we could curate the semantic annotations for as many as possible of these assays, and all the while refining our schema for how and what to annotate. The current interface looks something like this:
 Bioassay Express
Bioassay Express
The aesthetic design looked worse in the past, and will look better in the future, but the basic idea comes through: on the left is the text, which has been brought in from PubChem, to which it was submitted by the original scientist. On the right are a number of categories (derived from our common assay template), most of which has at least one assigned term. While the screenshot shown above displays these as short text labels, they are internally represented as URIs, and each of these links into a well defined ontology, with its own hierarchy and layers of semantic meaning (as per general principles of linked data).
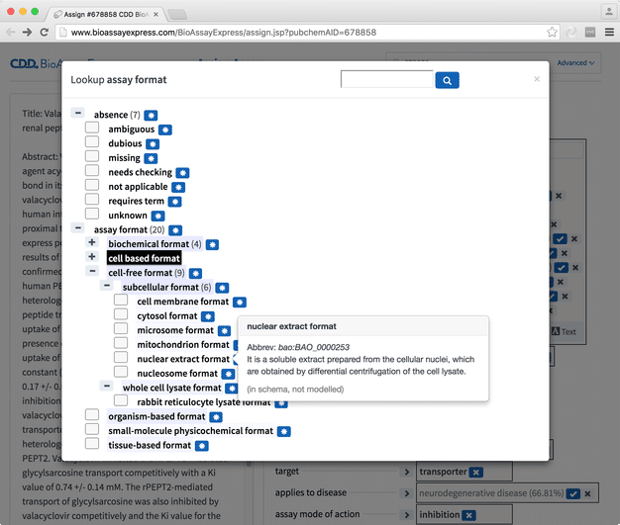
This is easier to see when a term is being selected using the Tree view dialog:
 Bioassay Express
Bioassay Express
There is quite a lot going on in the screenshot above, but the key piece of information is that all of the terms are represented in a hierarchy, and each of the labels comes with a lot more information than just the text.
Once we had the preliminary annotation interface working, our next task was to assemble a team of biologists to share their expertise by using the annotation system, which served several purposes: generating valuable training data, iteratively improving the schema, and of course testing the software. Fast forward to the present, and we have accumulated close to three thousand expert-curated assays (you can check out the current number anytime).
The reason why we are putting so much effort into representing bioassay protocols with semantic terminology is discussed in detail in our literature publications, but the long story short is that the current best practices for representing assays is to document them with plain text, so the best case scenario is that you will be able to access a summary description of half a page or so. If you want to compare two assays, and you are an expert in the field, and you have 10-15 minutes to carefully read both of them, you don’t need any help from the project we are working on. But if you want to compare many assays, or if you want to search a database using terms that are precise, your choices are unsatisfactory: searching is generally done by keyword searching, and any higher level analysis is done by direct machine learning from the text. With the appropriate semantic annotations, though, you can search for exactly what you want, with zero false positives, and zero false negatives.
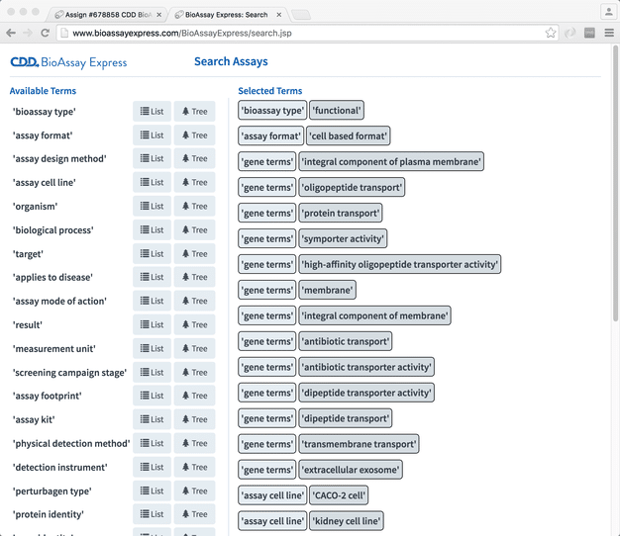
To demonstrate this, we have a preliminary search page:
 Bioassay Express
Bioassay Express
The way this works is basically to use all of the provided semantic terms (using an interface very similar to the annotation page) as fingerprints, in a way that is quite similar to comparing two molecules (e.g. using structure-derived fingerprints to compute a Tanimoto similarity metric). This allows anyone to pull out a list of assays from the database, sorted by most-similar-first.
This is just one demonstration of how assays can be located/selected using the annotations, rather than cruder methods (like keyword searching), but we are working on a variety of other techniques to allow people to hunt through a database of assays and hone in on the ones they want (if this sounds interesting, click on the Explore Assays page).
The project is evolving very rapidly, and the main users right now consist of just our team of biologists who are pushing it as far as (and further than) it was intended to go. One of the choices we made at the beginning was to operate the project out in the open. This is partly out of general principle (we are very pro-collaboration), but also convenience: we are a geographically distributed team, and the easiest way to deploy software to someone on the other side of the continent is to dump it onto a public website with no security. That is what you will find at http://www.bioassayexpress.com: the site is completely read-only at the moment, which means you don’t have to sign in, and you also can’t break anything. It is entirely possible to use it with your own assays and download annotated results, if you want to try it out.
As well as the openness of the website itself, we are very non-proprietary about the curated data that we are generating. We leveraged the PubChem service to get started, and all of the value that we are adding is available to anyone who wants it (there is a public API: it is nominally self-explanatory, to anyone who is a genuine übergeek). Part of the project source code is based on the open source project that we created for bioassay templates (see GitHub), but the main project that drives the website itself is proprietary. This is a for-profit venture, and as with many of the R&D projects that are carried out at Collaborative Drug Discovery, there is a dualism whereby the low level tools are made free and open to everyone, whereas the high level tools that bring everything together with maximum convenience cost money.


