From the desk of CDD VP Science, Sean Ekins, Ph.D., D.Sc.
At CDD, we are focused on novel strategies for developing inhibitors of Mycobacterium tuberculosis (Mtb) as tuberculosis (TB) drugs. Tuberculosis kills 1.8 million people every year (~1 every 8 seconds). Drug-drug interactions and co-morbidity with HIV are also important for TB. There has been a considerable increase in high throughput screening to identify hits as starting points for potential therapeutics. Even though 1000’s of hits have been identified (1-3) in this way we are lacking target assignments which would help in assessing utility and for target-based optimization.
We have therefore collated information on >700 molecules screened versus Mtb and their targets (4). This data is only available in Collaborative Drug Discovery, Inc. (CDD). We have added published data on target, essentiality, links to literature (PubMed), genes (tbdb.org), pathways (TBCyc, which provides a pathway-based visualization of the entire cellular biochemical network) and human homolog information.
We have now used this dataset and created an iOS app called TB Mobile that displays molecule structure and links to the bioinformatics data. By input of a molecule structure and performing a similarity search we can infer potential targets or search by targets to retrieve compounds known to be active. The app also has filters to limit the visible molecules by target name, pathway name, essentiality and human ortholog. Each molecule can be copied to the clipboard then opened with other apps (e.g. MMDS, MolPrime, MolSync, ChemSpider, and from these exported via Twitter or email) or shared via Dropbox.
TB Mobile is an example of appification of data and could be expanded to other drug discovery datasets. We will regularly update the dataset in TB Mobile so that it continues to be relevant.
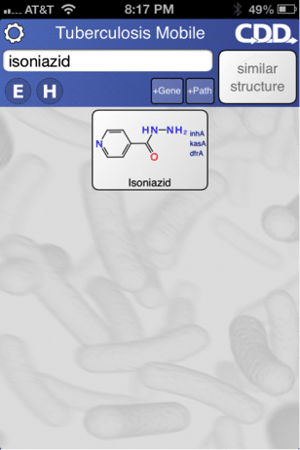
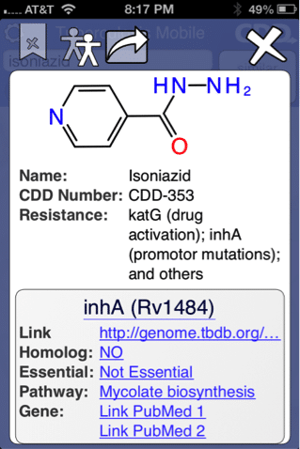
The app can be used to search for molecules e.g. the frontline drug isoniazid (above left). Molecule information and links out can viewed, shared or copied to the clipboard (above right).
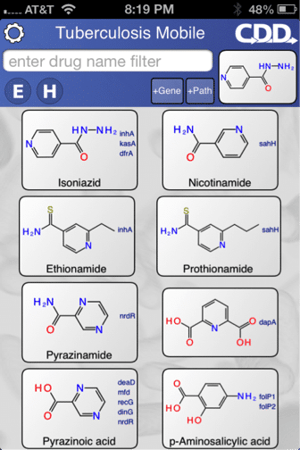
Similarity searching within the app enables ranking of other molecules active against TB and inference of potential targets.
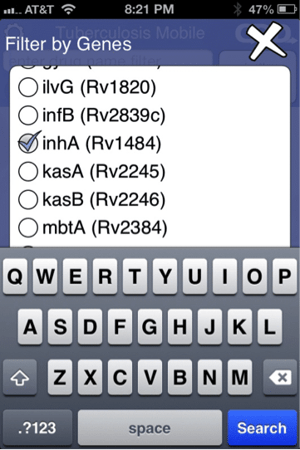
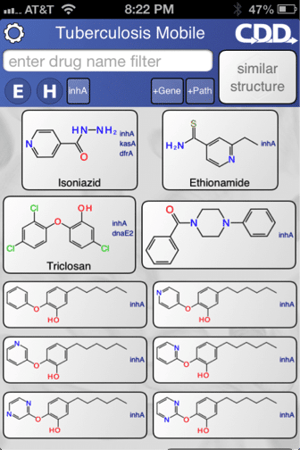
Data can also be filtered by target name, pathway name, essentiality and human ortholog (above left). For example, the image on the right shows the data filtered by the target InhA.
Credits
Alex M. Clark (Molecular Materials Informatics, Inc.) was responsible for app development, Malabika Sarker (SRI) and Sean Ekins (CDD) curated the data while the app was conceived of by Sean Ekins. Funding for the app comes from Award Number 2R42AI088893-02 “Identification of novel therapeutics for tuberculosis combining cheminformatics, diverse databases and logic based pathway analysis” from the National Institute of Allergy And Infectious Diseases. (PI: S. Ekins)
References
- Maddry, J. A.; Ananthan, S.; Goldman, R. C.; Hobrath, J. V.; Kwong, C. D.; Maddox, C.; Rasmussen, L.; Reynolds, R. C.; Secrist, J. A., 3rd; Sosa, M. I.; White, E. L.; Zhang, W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 2009, 89, 354-363.
- Ananthan, S.; Faaleolea, E. R.; Goldman, R. C.; Hobrath, J. V.; Kwong, C. D.; Laughon, B. E.; Maddry, J. A.; Mehta, A.; Rasmussen, L.; Reynolds, R. C.; Secrist, J. A., 3rd; Shindo, N.; Showe, D. N.; Sosa, M. I.; Suling, W. J.; White, E. L. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009, 89, 334-353.
- Reynolds, R. C.; Ananthan, S.; Faaleolea, E.; Hobrath, J. V.; Kwong, C. D.; Maddox, C.; Rasmussen, L.; Sosa, M. I.; Thammasuvimol, E.; White, E. L.; Zhang, W.; Secrist, J. A., 3rd. High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2011.
- Sarker, M.; Talcott, C.; Madrid, P.; Chopra, S.; Bunin, B. A.; Lamichhane, G.; Freundlich, J. S.; Ekins, S. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium Tuberculosis. Pharm Res 2012, 29, 2115-2127.
This blog is authored by members of the CDD Vault community. CDD Vault is a hosted drug discovery informatics platform that securely manages both private and external biological and chemical data. It provides core functionality including chemical registration, structure activity relationship, chemical inventory, and electronic lab notebook capabilities!
CDD Vault: Drug Discovery Informatics your whole project team will embrace!
Other posts you might be interested in
View All Posts
News
2 min
November 3, 2014
CDD Announces Partnership With Ryoka Systems
Read More
CDD Vault Updates
2 min
July 28, 2023
CDD Vault Update (July 2023 #2): Insert Abbreviations and Rotate Structures, Formatting Options in ELN, Move Batches via API, Share CDD Visualization
Read More
CDD Vault Updates
2 min
March 13, 2024
CDD Vault Update (March 2024): Redesigned Dose Response Viewer and a New GET Status (Background) API Call
Read More



