What is IC50 in the First Place?
An IC50 measurement tells us the concentration at which a drug is able to inhibit a particular biological process by 50%. For example, if compound A can inhibit 50% of the binding of a ligand to a particular receptor at a concentration of 5 nM, it would have an IC50 value of 5 nM. A closely related concept is the EC50 value which is the potency of a drug to induce a biological process.
So then what is a pIC50? It’s simply the negative log of the IC50 value in molar. Watch:
- An IC50 of 1 µM is 10-6 M, which is pIC50 = 6.0
- An IC50 of 1 nM is 10-9 M, which is pIC50 = 9.0
- An IC50 of 10 nM is 10-8 M, which is pIC50 = 8.0
- An IC50 of 100 nM is 10-7 M, which is pIC50 = 7.0
- An IC50 of 30 nM is 3 x10-8 M, which is also 10-7.5 M, which is pIC50 = 7.5
Do you see a pattern? You’re in drug discovery… you’ve been working with pH since you were knocking over graduated cylinders in high school. When working in the lab, did you say that this solution has an acidity of 10 µM (10-5 M) [H+]? Of course not. You talked about a solution at pH 5. And you didn’t bother trying to back-calculate in your head … that is, you didn’t try and convert a pH value of 7.5 to 30 nM [H+]. That’s because you’ve trained yourself to think in terms of pH, as well as the fact that the acidity of an aqueous solution is a logarithmic function. pH values in experiments go from 1 to 2 to 3, not 10 mM to 20 mM to 30 mM [H+].
That is exactly the way you should think about IC50 values (or when testing agonists, EC50 values). Dose-dependent inhibition (or activation) of an enzyme or cell is a logarithmic phenomenon (with regard to compound concentration), so it makes more sense to view the data this way. And if you look at papers from the big pharma companies, you will notice that they more often than not report inhibition as pIC50 instead of IC50.
So should you do it that way just because Merck does it that way? No, so let’s discuss why it is the right way (and why Merck, GSK, etc. use it).
The Dose-Response Curve
When you crunch your dose response data, you are fitting your response (say 0-100%, to keep it simple) against the concentration of test compound (ultimately in molar). But if you look carefully at the equation for which you are running a curve fit (the Four-Parameter Logistic Function, also known as the sigmoid function), what is being calculated is response vs. the log of compound concentration.
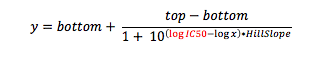
where y is the response you have measured at compound concentration x. Top is the response with no inhibitor (aka the maximum asymptote), and bottom is the response when the enzyme (or cell) is fully inhibited (aka the minimum asymptote). And of course, IC50 is the inflection point (halfway point between top and bottom), the value you are solving for.
So the curve fitting actually solves for the logIC50, not IC50. If you want to know the error from the fit, you can get a standard error (SEM) that surrounds the IC50 symmetrically. But if you start changing the pIC50 value to IC50, that error becomes asymmetric, and makes no sense.
You have to go back and do the anti-log to get the IC50. And this is easy these days with computers and Excel. So “everyone does it”. But you lose a lot in taking that “easy step”. What is the advantage? How will this change your life?